研究方向
Comparative genomics
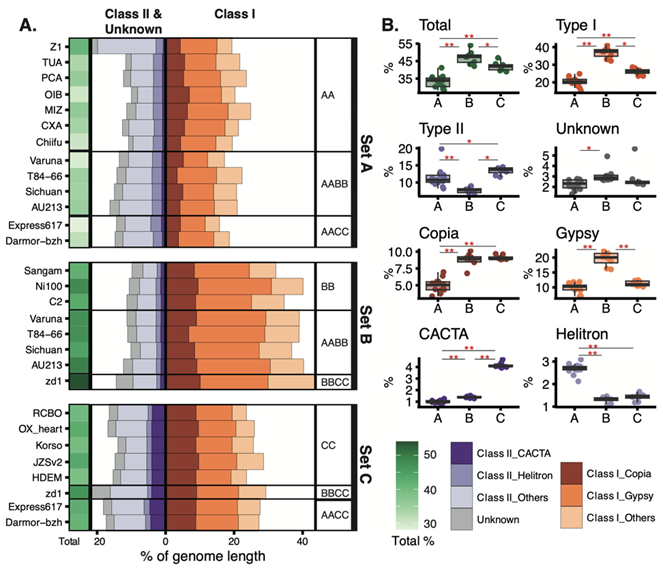
We are generally interested in analyzing complex genomes to understand the changes and causes of genome structures, either driven by evolution, domestication, or engineering, including species in bacteria, fungi, diatoms, plants, and humans. In addition to genome assembly, gene prediction, and comparative genome analysis, we are developing and incorporating new computational and genomic tools to analyze genomic information. High-throughput sequencing data (Illumina, PacBio, and Oxford Nanopore) with chromosome conformation capture (Hi-C) data were frequently combined in our analysis to detect complex genome structures.
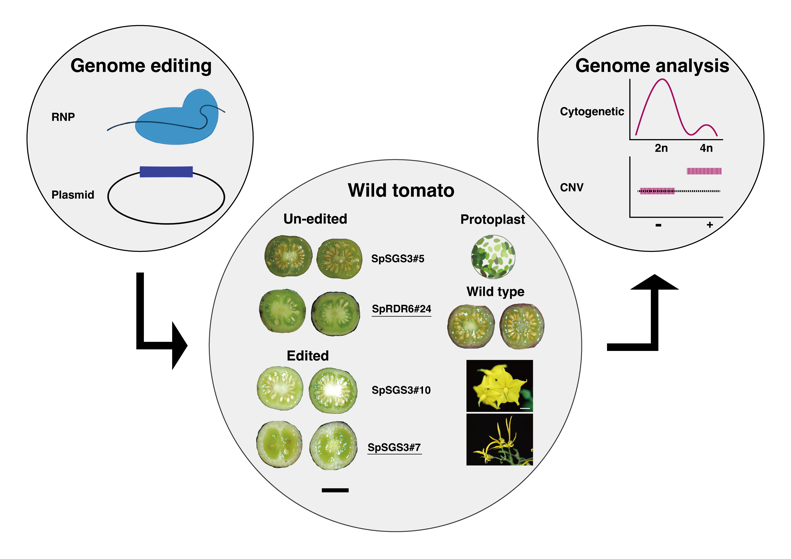
Tissue regeneration, genome engineering, and manipulation are routinely performed in functional studies, pharmaceutical production, and plant breeding. However, the effect of these cellular stresses on genome stability remains unknown. We developed a novel bioinformatics analysis using state-of-the-art sequencing methods to understand the somatic mutation spectrum, copy number variations, and abnormal chromosomal changes in human cell lines and polyploid plant genomes.An in-house high-performance computing (HPC) cluster is the powerhouse for analyzing genomic data.
Systems Genetics of Crop Stress Biology
A tightly regulated complex signal transduction network and chaperone machinery controls gene expression and enables the cell to change its transcriptional capacity within minutes in the presence of stress. Multiple genes and regulatory networks have been identified that are involved in heat or flooding responses in different plants. Plant reproductive development is particularly vulnerable to stressful conditions and causes substantial yield losses. The goal of this research is to integrate association mapping and genomic selection to identify changes in gene expression levels, in particular alternative splicing (AS), allele specific expression (ASE) and transposable elements (TEs), which are involved in responses to abiotic stress.
Plant Phenotyping
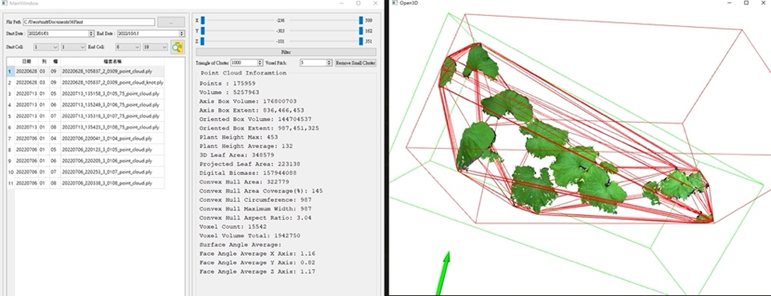
The imaging base, a non-invasive plant phenotyping system, can routinely measure plant phenotypes in a high-throughput manner. However, the connections between plant phenotypes and internal phenotypic states, including cellular, tissue, and physiological properties, remain unclear. Little information regarding the transcriptional, translational, and physiological states of plants can be obtained using the external phenotype. We are currently developing a crop physiological phenotyping platform using a holistic multidimensional omic approach. Machine vision with advanced statistics and machine learning frameworks will be developed to process imaging-based traits. This will allow us to select imaging features that can act as a proxy for the underlying physiological processes.

葉國楨
特聘研究員兼主任
主任(2019.1.16 至今)
代理主任 (2016.10.1-2019.1.15)
學程召集人 中研院國際研究生院 分子及生物農業科學 博士班學程(2015-2020)
特聘研究員(2024 至今)
研究員 (2014 -2024)
副研究員 (2009-2013)
助研究員 (2001-2009)
博士後研究 Howard Hughes Medical Institute and Department of Biological Sciences, Stanford University, USA (1999-2001)
博士 Plant Biology Graduate Group, University of California, Davis, USA (1994-1999)

